(h2) Stereoscopic Height Measurement of Point-like Features
For point-like features (for instance ``moss'' or small-scale loops of
the size of one or a few pixels) that are visible in both the STEREO A
and B images), the height can be measured from the stereoscopic parallax.
The routine EUVI_CORR.PRO allows the user to click on such a structure in
image A and B, and then calculates the stereoscopic height from the centroid
determined in a 3x3 pixel neighborhood of the points.
Let us assume the user has produced a coaligned pair of stereoscopic images
in array IMAGE_PAIR[2, NX, NY] and the image parameters are in the
variable PARA, both stored in the savefile 'loop_A.sav'. We will store the
3D coordinates of the moss feature in the file 'moss_A.dat':
IDL>
savefile='loop_A.sav'
restore,savefile
;restore saved data from step (2a)
nsm =3
;smoothing boxcar for highpass filter
fov_moss=[710,850,750,910]
;field-of-view in pixels [i1,j1,i2,j2]
dcont =5
;contour level step for logarithmic contours LEVEL=DCONT * 2^N
mossfile='moss_A.dat'
;output filename where 3D coordinates of loops are stored
ct =3
;contour level step for logarithmic contours LEVEL=DCONT * 2^N
euvi_corr,image_pair,para,nsm,fov_moss,dcont,mossfile,ct
The output on the screen will look like this:

After the user clicked a feature in image A (right), the estimated position
is projected onto image B (left), with a cross, and the user can click on
the peak location of the corresponding feature in image B. The program
determines the centroids of the 3x3 pixel neighborhoods (marked with red squares)
by a parabolic fit (see display with parabolic fit in window 1).
The output in window 1 will look like this:

The calculated
height is then displayed and the user can decide to save this value in the
datafile "moss_A.dat". The procedure can be repeated for an arbitrary number
of features, which can subsequently be appended to the same datafile "moss_A.dat".
Let us assume you have identified 30 moss features and saved the data in the
file "moss_A.dat", which could look like this:
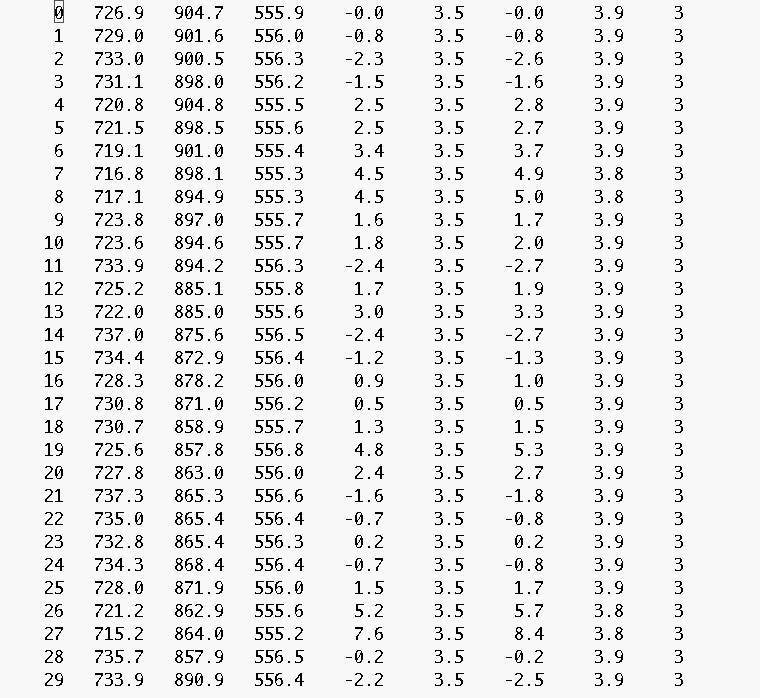
In order to create a summary plot in the context of the entire FOV of the
active region we can call:
IDL>
fov =[640,820,800,980]
;field-of-view in pixels [i1,j1,i2,j2]
euvi_corrplot,image_pair,para,nsm,fov_moss,dcont,mossfile,ct,fov
The output on the screen will look like this:

A postscript file will be created with the name corrplot_a_col.ps



